Preliminary notes on synthetic control
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
data = pd.read_csv('../data/cigs.csv')
data
| state | year | cigs | loggdppercap | retprice | pctage1524 | beerpercap | cigsalespercap88 | cigsalespercap80 | cigsalespercap75 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | AL | 1970 | 89.8 | 9.679 | 89.344 | 0.175 | 18.96 | 112.1 | 123.2 | 111.7 |
| 1 | AL | 1971 | 95.4 | 9.679 | 89.344 | 0.175 | 18.96 | 112.1 | 123.2 | 111.7 |
| 2 | AL | 1972 | 101.1 | 9.679 | 89.344 | 0.175 | 18.96 | 112.1 | 123.2 | 111.7 |
| 3 | AL | 1973 | 102.9 | 9.679 | 89.344 | 0.175 | 18.96 | 112.1 | 123.2 | 111.7 |
| 4 | AL | 1974 | 108.2 | 9.679 | 89.344 | 0.175 | 18.96 | 112.1 | 123.2 | 111.7 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1204 | WY | 1996 | 110.3 | 9.914 | 81.000 | 0.174 | 24.98 | 114.3 | 158.1 | 160.7 |
| 1205 | WY | 1997 | 108.8 | 9.914 | 81.000 | 0.174 | 24.98 | 114.3 | 158.1 | 160.7 |
| 1206 | WY | 1998 | 102.9 | 9.914 | 81.000 | 0.174 | 24.98 | 114.3 | 158.1 | 160.7 |
| 1207 | WY | 1999 | 104.8 | 9.914 | 81.000 | 0.174 | 24.98 | 114.3 | 158.1 | 160.7 |
| 1208 | WY | 2000 | 90.5 | 9.914 | 81.000 | 0.174 | 24.98 | 114.3 | 158.1 | 160.7 |
1209 rows × 10 columns
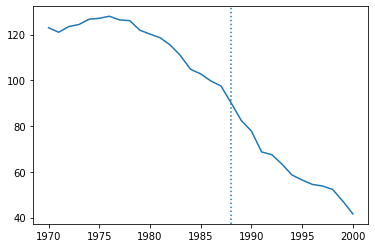
dcal = data.query("state=='CA'")
plt.plot(dcal['year'],dcal['cigs'])
plt.axvline(1988,linestyle=":")
plt.show()
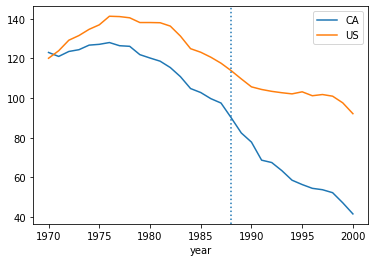
dcal = data.query("state=='CA'")
plt.plot(dcal['year'],dcal['cigs'],label="CA")
data.query("state != 'CA'").groupby('year')['cigs'].mean().plot(label="US")
plt.axvline(1988,linestyle=":")
plt.legend()
plt.show()
Checking pretrends
We can regress using the other states as control.
data['D'] = (data['state']=='CA') & (data['year']>=1988)
data['T'] = (data['state']=='CA')
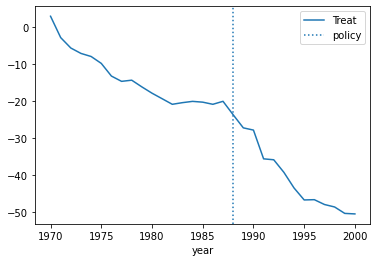
To evaluate the pre-trend, we can estimate the coffecient of the interaction with the values leading to the event.
import statsmodels.formula.api as smf
# Initialise and fit linear regression model using `statsmodels`
model = smf.ols("cigs ~ C(year) + C(year):C(T)", data=data)
model = model.fit()
data_cal = data.query("state == 'CA'").copy()
data_cal['dep'] = model.predict(data_cal) - model.predict( data_cal.eval("T=False") )
data_cal.plot(x='year',y='dep',label="Treat")
plt.axvline(x=1988, linestyle=':',label="policy")
plt.legend()
<matplotlib.legend.Legend at 0x7f60f1d68d90>
Construct non treated outcome using observables
In this section we think about directly constructing a control group as a weighted sum of available non treated units. A simple choice would be to pick the state with the most similar observables. Or we could weight becased on the distance on obserbales.
Let's try both of these approaches.
Next we might want to change the way we construct our non-treated counterfactual for California. First we could choose to use observables characteristics to be added to our framework.
loggdppercap retprice pctage1524 beerpercap
# we start by computing pair-wise distances between observations